Robustness: sensitivity ethnic names
Thijmen Jeroense
Last compiled on 26 februari, 2024
Preparing data
Libraries
fpackage.check <- function(packages) { # (c) Jochem Tolsma
lapply(packages, FUN = function(x) {
if (!require(x, character.only = TRUE)) {
install.packages(x, dependencies = TRUE)
library(x, character.only = TRUE)
}
})
}
packages = c("haven", "NSUM", "coda", "matrixStats", "parallel",
"doParallel", "tictoc", "beepr", "tidyverse",
"stringr", "viridis", "kableExtra","ggridges", "viridis", "ggdark", "ggplot2", "patchwork", "sjPlot")
fpackage.check(packages)Prepare and execute NSUM models
Preparing ethnic names.
To assess the sensitivity of network size estimates we estimate the main NSUM estimation, with prison as unknown population, with different ethnic name combinations. So first we have to prepare the model specifications for each of these ethnic name combinations and store them in a list to use in the iteration.
load(file = "data_analysis/data/data_processed/nells_data/2022-11-09_nells-nsum-prepped-data.rds")
frequencies <- read_csv("data_analysis/2022-08-26_namefrequencies.csv")
#------------------------------------------------------------------------------------------------
nells_nsum <- nells_nsum %>%
arrange(id)
#-- create 10 different options.
mat_list <- list()
# All ethnic names
mat_list[[1]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_mohammed_boundary,
knows_fatima_boundary,
knows_ibrahim_boundary,
knows_esra_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list <- list()
known_list[[1]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_mohammed",
"knows_fatima",
"knows_ibrahim",
"knows_esra",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# ibrahim
mat_list[[2]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_ibrahim_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[2]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_ibrahim",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# esra
mat_list[[3]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_esra_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[3]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_esra",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# fatima
mat_list[[4]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_fatima_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[4]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_fatima",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# fatima and mohammed
mat_list[[5]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_mohammed_boundary,
knows_fatima_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[5]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_mohammed",
"knows_fatima",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# esra and ibrahim
mat_list[[6]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_ibrahim_boundary,
knows_esra_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[6]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_ibrahim",
"knows_esra",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# mohammed and ibrahim
mat_list[[7]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_ibrahim_boundary,
knows_mohammed_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[7]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_ibrahim",
"knows_mohammed",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# esra and fatima
mat_list[[8]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_esra_boundary,
knows_fatima_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[8]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_esra",
"knows_fatima",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# esra and mohammed
mat_list[[9]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_esra_boundary,
knows_mohammed_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[9]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_esra",
"knows_mohammed",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
# fatima and ibrahim
mat_list[[10]] <- nells_nsum %>%
dplyr::select(knows_daan_boundary,
knows_kevin_boundary,
knows_edwin_boundary,
knows_albert_boundary,
knows_emma_boundary,
knows_linda_boundary,
knows_ingrid_boundary,
knows_willemina_boundary,
knows_fatima_boundary,
knows_ibrahim_boundary,
knows_prison_boundary,
knows_mbo,
knows_hbo,
knows_university,
knows_secundary,
knows_secondhome,
knows_unemployed) %>%
as.matrix()
known_list[[10]] <- frequencies %>%
filter(
name %in% c(
"knows_daan",
"knows_kevin",
"knows_edwin",
"knows_albert",
"knows_emma",
"knows_linda",
"knows_ingrid",
"knows_willemina",
"knows_fatima",
"knows_ibrahim",
"knows_prison",
"knows_mbo",
"knows_hbo",
"knows_university",
"knows_secundary",
"knows_secondhome",
"knows_unemployed"
)
) %>%
pull(number)
unknown <- c(13,10,10,10,11,11,11,11,11,11)NSUM estimation
Estimate the different NSUM models.
#------------------------------------------------------------------------------------------------
# paralellize the estimation
numCores <- detectCores()
# so we need to do in seperate runs
registerDoParallel(core=numCores-1)
######################################################
#NSUM analysis
######################################################
#set seed
set.seed(220914)
#------------------------------------------------------------------------------------------------
# some info to work with
iters <- 40000 # number of iterations (40k)
burns <- 1000 # burnin size (1k)
retain <- 4000 # how many chains do we want to retain? (4k)
popsize <- 17407585 # population size
# fill up empty lists
kds <- list()
kdssd <- list()
data_list <- list()
#------------------------------------------------------------------------------------------------
foreach(i = 8:10) %dopar% {
# paralellize the estimation# i = 1
# 8 cores in total so we can use 7 cores for the parallel computing.
cd <-
mat_list[[i]][,-c(unknown[i])] # take out pop of interest and make a new mat
file_name_data <-
paste0(
"data_analysis/results/nsum_output/robustness/ethnic_names/data/estimates_holdout",
i,
".txt"
) #create file.name for data
file_name_model <-
paste0(
"data_analysis/results/nsum_output/robustness/ethnic_names/model/estimates_holdout",
i,
".rds"
) #create file.name for model
if (!file.exists(file_name_model)) {
# calculate starting values
z <-
NSUM::killworth.start(cbind(cd, mat_list[[i]][, c(unknown[i])]), # paste the "takenout" at the END of matrix
known_list[[i]][-c(unknown[i])], # this is the known pop, WITHOUT unknown pop
popsize) # population size
degree <-
NSUM::nsum.mcmc(
cbind(cd, mat_list[[i]][, c(unknown[i])]),
# gets pasted at the last column
known_list[[i]][-c(unknown[[i]])],
# here we again take out the "holdout", or artificial "unknown" pop
popsize,
model = "combined",
# combined control for transmission and recall errors
indices.k = ncol(mat_list[[i]]),
# notice that "holdout" gets pasted as last column
iterations = iters,
burnin = burns,
size = retain,
# 40k iterations, retain 4k chains
d.start = z$d.start,
mu.start = z$mu.start,
# starting values from simple estimator
sigma.start = z$sigma.start,
NK.start = z$NK.start
)
save(degree, file = file_name_model)
} else {
load(file_name_model)
}
#store mean and sd in df
# calculate rowmean and it's SD of the retained 4k chains
kds[[i]] <-
rowMeans(degree$d.values, na.rm = TRUE) # calculate rowmean of netsize iterations: so the retained chains
kdssd[[i]] <-
matrixStats::rowSds(degree$d.values) # calculate sd of 4k estimates per row: sd for those values
data_list[[i]] <-
data.frame(cbind(kds[[i]], kdssd[[i]])) # combine and put in df
# Save the data, new .txt for each iteration, if something goes wrong, we can always start at prior one and combine
write.table(data_list[[i]], file = file_name_data, row.names = F) #store them in results.
}Combine results
Combine the different results and combine with information from the NELLS.
#import nells file.
if (file.exists(
"data_analysis/results/nsum_output/robustness/ethnic_names/combined_data/df_models_nsum_long.rds"
)) {
load(file = "data_analysis/results/nsum_output/robustness/ethnic_names/combined_data/df_models_nsum_long.rds")
} else {
list_files <-
as.list(
dir(
"data_analysis/results/nsum_output/robustness/ethnic_names/model/",
full.names = T
)
)
#create loop lists
kds <- list()
kdssd <- list()
data <- list()
list_df <- list()
#loop to extract information
for (i in 1:length(list_files)) {
#i = 1
print(paste0("Number ", i, " of ", length(list_files)))
load(list_files[[i]])
kds[[i]] <-
rowMeans(degree$d.values, na.rm = TRUE) # calculate rowmean of netsize iterations: so the retained chains
kdssd[[i]] <-
matrixStats::rowSds(degree$d.values) # calculate sd of 4k estimates per row: sd for those values
data[[i]] <- cbind(kds[[i]], kdssd[[i]]) # combine them
list_df[[i]] <-
cbind(as_tibble(data[[i]]), nells_nsum$id) # add NELLS id variable
strings <-
str_split(str_extract(list_files[[i]][1], pattern = "estimates.+"),
pattern = "_") # add holdout and tauk number
list_df[[i]] <- list_df[[i]] %>%
mutate(
holdout = as.numeric(str_extract(strings[[1]][2], pattern = "[[:digit:]]{1,}")))
#combine results and save
df_models_nsum_long <- list_df %>%
bind_rows() %>%
rename(mean = V1,
sd = V2,
id = 3)
#save iamge
save(df_models_nsum_long, file = "data_analysis/results/nsum_output/robustness/ethnic_names/combined_data/df_models_nsum_long.rds")
}
}
#robustness data
df_robustness <- df_models_nsum_long
#load mohammed data
load("data_analysis/results/nsum_output/main/old/df_models_nsum_long.rds")
mohammed <- df_models_nsum_long %>%
filter(holdout == 10)
mohammed <- mohammed %>%
mutate(holdout = 11)
mohammed <- nells_nsum %>%
dplyr::select(id, migration_background_fac, gender) %>%
right_join(mohammed)
test_df <- nells_nsum %>%
dplyr::select(id, migration_background_fac, gender) %>%
right_join(df_robustness) %>%
bind_rows(mohammed) %>%
mutate(spec_ethnic = factor(
holdout,
levels = 1:11,
labels = c(
"All",
"Ibrahim",
"Esra",
"Fatima",
"Fatima_Mohammed",
"Esra_ibrahim",
"Ibrahim_Mohammed",
"Esra_Fatima",
"Esra_Mohammed",
"Fatima_Ibrahim",
"Mohammed"
)))Sensitivity analysis
We want to know how sensitive the extended network size is to inclusion and exclusion of different ethnic names. To check this we thus specified different models (with same unknown population: prison) and we can check how different estimations are within groups.
The first difference we need to check is the difference between a single name and multiple names. Since the names differ by gender they are are related to we also check gender differences within groups.
First of all, figure 1 shows that including all names severely increases the estimated extended network size of the minoritised groups. This is not that surprising as using all ethnic names means 33% of names is used to estimate 8% of the population.
Second, given that gender homophily preferences are high, we would ideally want to have at least an ethnic male and female name. However, In this situation we would still over-represent the ethnic minorised by a factor of 2 (16.5%/8%), which is also reflected in the extended network size estimates. Hence our decision to use only one ethnic name.
Even though overall size estimates change substantially when we included different name combinations, the differences between male and female members of groups seems to be relatively stable. However, we need to take a closer look to choose the name for final analysis.
test_df %>%
filter(gender != "Other") %>%
filter(spec_ethnic %in% c("Ibrahim",
"Esra",
"Fatima",
"Mohammed",
"Esra_Fatima",
"Ibrahim_Mohammed",
"Esra_Fatima",
"Esra_Mohammed",
"Fatima_Ibrahim",
"All")) %>%
ggplot(aes(x = mean,
y = spec_ethnic,
fill = gender,
colour = gender
)) +
geom_boxplot(alpha = 0.4) +
facet_wrap(vars(migration_background_fac)) +
theme_minimal() +
scale_fill_viridis_d(option = "D") +
scale_color_viridis_d(option = "D") +
theme(axis.title.y = element_text(hjust = 1),
axis.title.x = element_text(hjust = 1),
text = element_text(colour = "black"),
strip.text = element_text(colour = "black")) +
labs(x = "Extended network size",
y = "Ethnic names included",
title = "Fig 1. Group and gender sensitivity in network size estimates to varying ethnic names")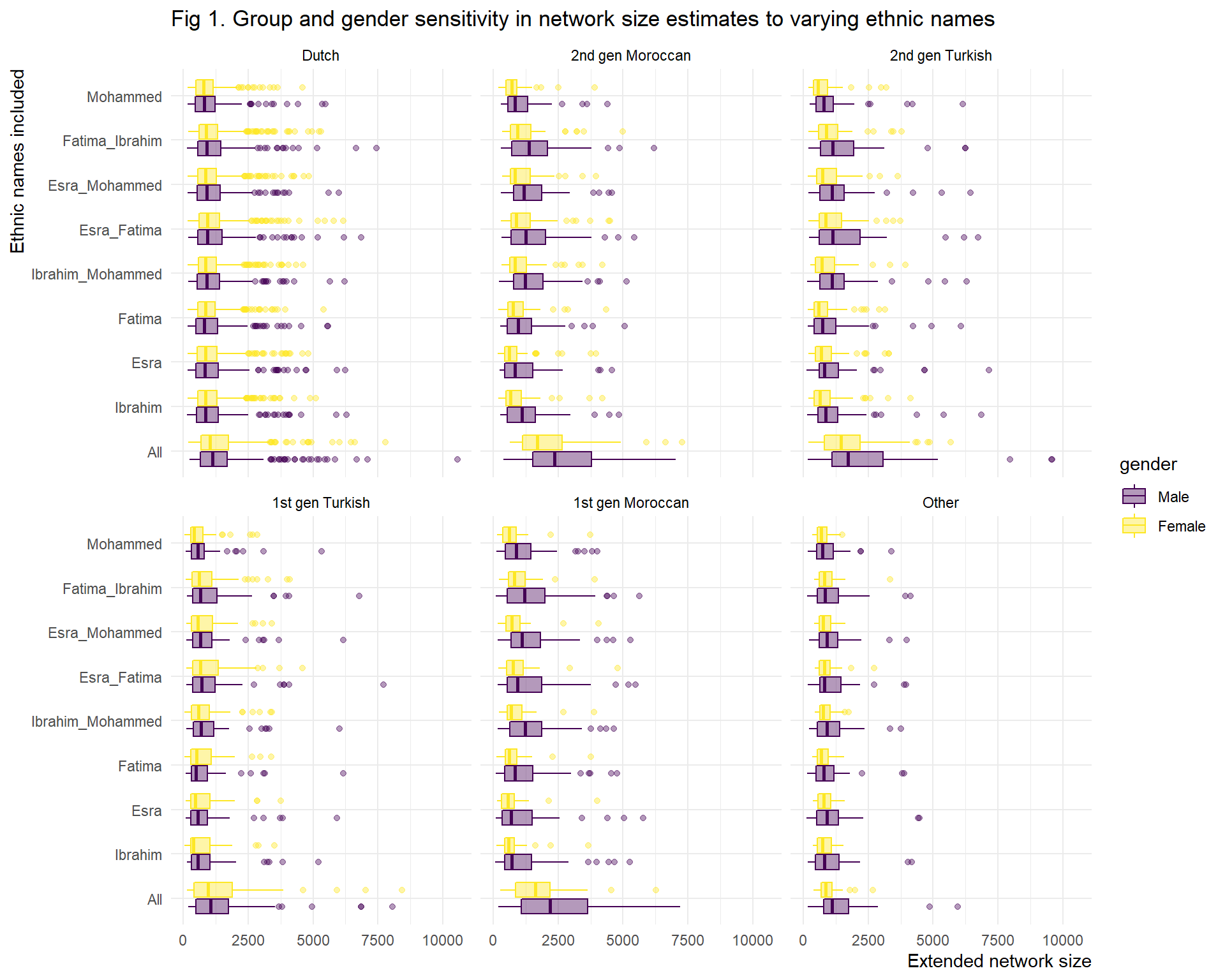
Given the decision to use one of the four ethnic names, we need to take a closer look at differences within and between these groups for individual names (fig. 2). There are some things to note. First of all, migrant extended networks are greater when an ethnic name that is affiliated with their group is used. This difference is greater for first generation than for second generation migrants. Moreover, the 1st gen Moroccan-Dutch are most sensitive to this bias. Second, there are some gender effects. Overall the gender difference remain the same within the group when we include different names, males know more than females, even when we use female names. There is one exception, namely the gender gap within 1st gen Turkish-Dutch, when we use Fatima as ethnic name. So we need to select the name with the least gender, migration generation and migration background bias.
test_df %>%
filter(gender != "Other") %>%
filter(spec_ethnic %in% c("Ibrahim",
"Esra",
"Fatima",
"Mohammed")) %>%
ggplot(aes(x = mean,
y = spec_ethnic,
fill = gender,
colour = gender
)) +
geom_boxplot(alpha = 0.4) +
facet_wrap(vars(migration_background_fac)) +
theme_minimal() +
scale_fill_viridis_d(option = "D") +
scale_color_viridis_d(option = "D") +
theme(axis.title.y = element_text(hjust = 1),
axis.title.x = element_text(hjust = 1),
text = element_text(colour = "black"),
strip.text = element_text(colour = "black")) +
labs(x = "Extended network size",
y = "Ethnic names included",
title = "Fig 2. Group and gender sensitivity in network size estimates to varying ethnic names")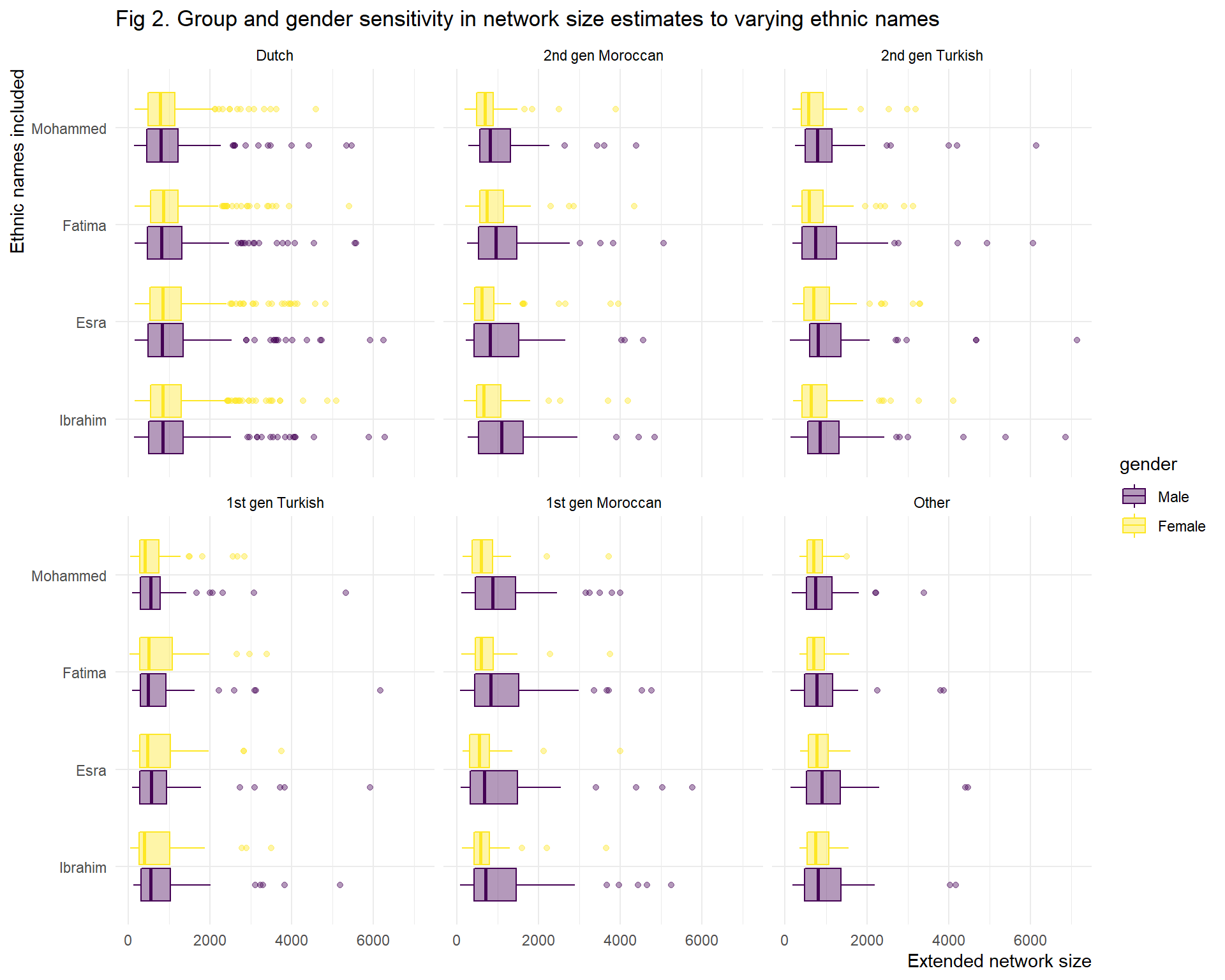 The generation and group sensitivity to different names is also apparent
when we look at the prevalence to know someone with a given name. We do
so by estimating different negative binomial regressions with only group
membership as independent variable.
The generation and group sensitivity to different names is also apparent
when we look at the prevalence to know someone with a given name. We do
so by estimating different negative binomial regressions with only group
membership as independent variable.
# use a loop.
#set var_names to use in loop.
variable_names_model <- c(
"knows_mohammed",
"knows_fatima",
"knows_ibrahim",
"knows_esra")
#start analsis loop
model_results <- list()
for(i in 1:length(variable_names_model)) {#i = 1
fm <- as.formula(paste(variable_names_model[[i]], "~", "migration_background_fac"))
model_results[[i]] <- MASS::glm.nb(fm,
data = nells_nsum)
}
#clean output with tidy r
model_results <- model_results %>%
purrr::map(.x =.,
.f = ~ broom::tidy(.x))
#add var_names to model_results
for(i in 1:length(model_results)){
model_results[[i]] <- model_results[[i]] %>%
mutate(dep_var = variable_names_model[i])
}
#combine model dfs.
model_results_df <- model_results %>%
bind_rows()
#set correct variable names
model_results_df <- model_results_df %>%
mutate(
term = case_when(
str_detect(term, "2nd gen Moroccan") ~ "2nd gen Moroccan",
str_detect(term, "2nd gen Turkish") ~ "2nd gen Turkish",
str_detect(term, "1st gen Moroccan") ~ "1st gen Moroccan",
str_detect(term, "1st gen Turkish") ~ "1st gen Turkish",
str_detect(term, "Other") ~ "Other",
term == "(Intercept)" ~ "Intercept"
)
)
#Set correct names
correct_names <- model_results_df %>%
pull(dep_var) %>%
str_replace(., pattern = "knows_", replacement = "") %>%
str_to_title()
#drop old names and add the correct names
model_results_df <- model_results_df %>%
dplyr::select(-dep_var) %>%
mutate(dep_var = correct_names)The results of this analysis are presented in figure 3. These show that the group difference in knowing someone is smallest for Ibrahim. Both the generation and the group gap are smallest when we use this name in the NSUM estimation, compared to Fatima, Mohammed, and Esra. This is why we have chosen to use Ibrahim in the final analysis.
#plot for ethnic names
model_results_df %>%
filter(term != "Intercept") %>%
ggplot(aes(x = dep_var,
y = estimate,
shape = term)) +
geom_linerange(aes(ymin = estimate - (std.error *1.96),
ymax = estimate + (std.error *1.96)),
position = position_dodge(width = 1)) +
geom_point(position = position_dodge(width = 1),
aes(colour = term,
fill = term)) +
geom_hline(yintercept = 0,
colour = "black") +
facet_wrap(vars(dep_var),
scales = "free_x",
ncol = 4) +
scale_fill_viridis_d(option = "E") +
scale_color_viridis_d(option = "E") +
scale_shape_manual(values = c(21,22,23,24,25)) +
scale_y_continuous(limits = c(-2,2.5)) +
theme_minimal() +
theme(#legend.position = 'none',
#plot.background = element_rect(fill = '#404040', colour = '#404040'),
#panel.background = element_rect(fill = '#404040', colour = '#404040'),
axis.title.y = element_text(hjust = 1),
axis.title.x = element_text(hjust = 1),
text = element_text(colour = "black"),
strip.text = element_text(colour = "black"),
axis.text.x = element_blank()) +
labs(x = "",
y = "B",
colour = "",
shape = "",
fill = "",
title = "Fig 3. Group differences in prevalence of knowing someone")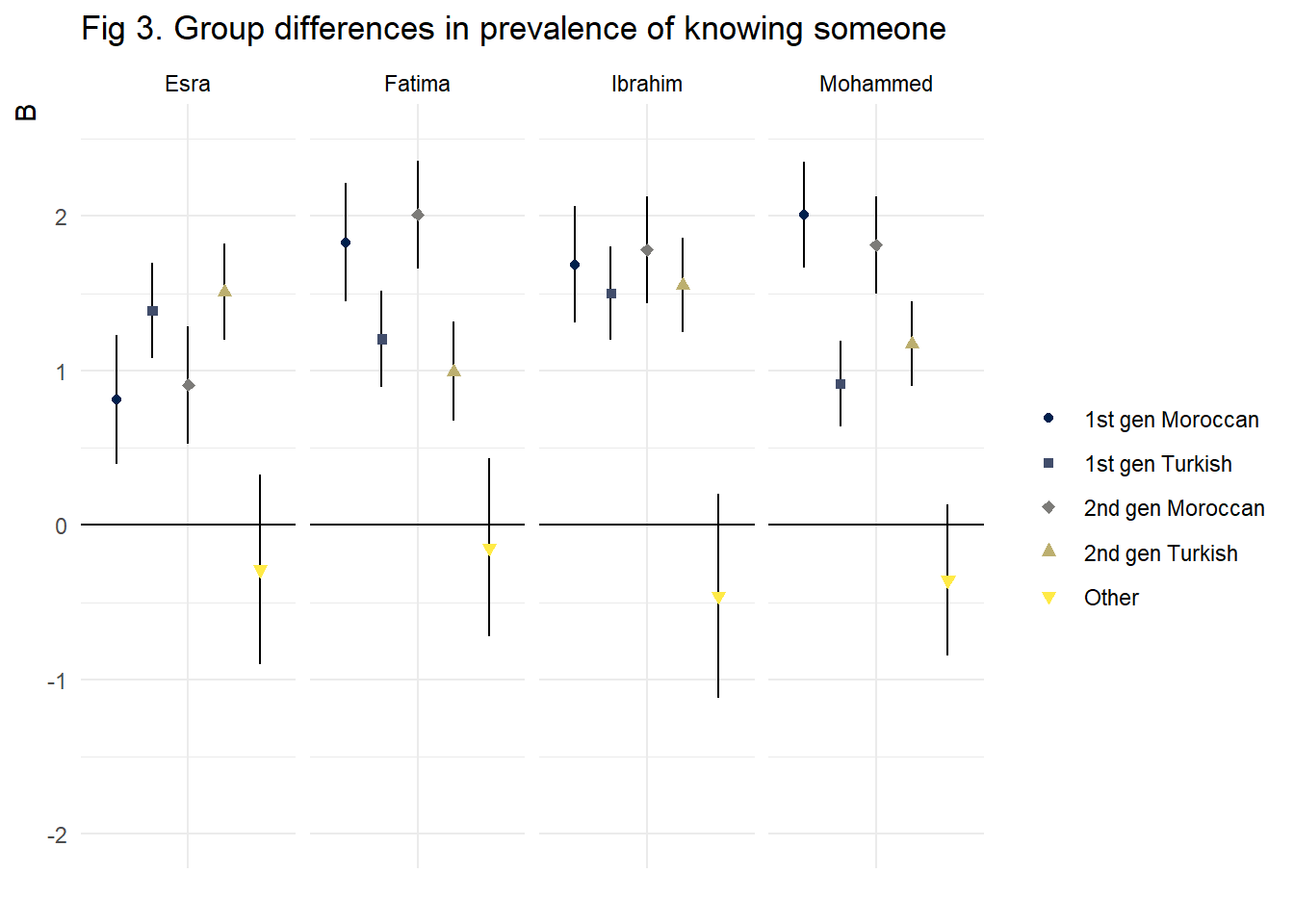
Copyright © 2024 Jeroense Thijmen